By Moiz Kohari, VP of Enterprise & Cloud Programs
Today, there is no application of artificial intelligence (AI) more consequential to human life than its use in healthcare and life sciences. The emotional toll of a life-threatening diagnosis — the uncertainty, the helplessness, the heartbreak — is something no family should have to endure. Our mission is not just to build systems; it is to accelerate the discovery of cures and to create an infrastructure that helps save lives. I hope the work my team and I share here can serve as a blueprint –– a reference architecture that healthcare institutions around the world can adopt and improve upon.
Unlike industries such as finance or pharmaceuticals, where competition often fuels secrecy around architectural breakthroughs, research hospitals should operate under a singular mandate: preserve and extend human life. Yes, hospitals compete fiercely — for talent, grants, patient outcomes — but that competition should live in the execution layer, not in the foundational systems. We should not hide the architectural blueprints that can help detect cancers earlier, reduce diagnostic error rates, or personalize treatment protocols more precisely. What Linux did for computing — establishing a common, open-source foundation — we must now do for data infrastructure in healthcare. We are building not just a solution, but a shared operating system for medical AI.
This article presents that blueprint. It details how modern AI techniques, from multimodal learning to federated inference, can reshape the diagnostic process, and help institutions detect disease with both speed and precision. Early detection is not just medically beneficial; it is statistically transformative. For example, when breast cancer is caught at Stage 1, the 5-year survival rate exceeds 99%, but that rate drops to 30% or less if caught at Stage 4 [ACS, 2023]. The challenge? A patient’s record is no longer a folder — it’s a high-dimensional system containing thousands of structured and unstructured data points: imaging, genomics, clinical notes, drug history, and more. AI is the only tool we have that can consume, contextualize, and learn from this complexity at scale.
Yet, with this power comes responsibility. Patient privacy is not just a legal requirement — it is an ethical obligation. As AI systems ingest and act on sensitive data, the risk of breach or misuse becomes existential. This is why federated AI must become a cornerstone of healthcare architecture. Models must learn collaboratively without centralizing sensitive data. By training AI across institutions without sharing the data itself, we preserve patient privacy while amplifying collective intelligence. This approach is already showing promise in early studies: one 2022 paper from Nature Medicine showed that federated AI improved cancer diagnosis rates across geographically distributed hospitals while maintaining compliance with GDPR and HIPAA frameworks.
In short: if we align our technical ambition with our shared humanity, we can build something that not only works — but endures. Something that no one hospital owns, but that every patient benefits from.
Why Federated AI Is Essential for Modern Healthcare
As hospitals, research institutions, and diagnostic labs begin to adopt AI at scale, one question becomes increasingly central: how can we harness the collective intelligence of distributed medical data without compromising privacy, control, or compliance?
The answer lies in Federated AI, a privacy-preserving framework that allows machine learning models to be trained across multiple institutions without the need to move or centralize patient data.
In traditional machine learning, data from multiple sources is pooled into a single repository to train a model. But in healthcare, this isn’t practical or even legal. Patient records are protected by regulations like HIPAA, GDPR, and a growing patchwork of regional and institutional governance policies. These rules exist for good reason: sensitive medical data can’t leave its originating facility without significant risk.
Federated AI flips the paradigm.
Instead of moving the data, federated learning sends the model to the data. Local models are trained within each institution’s secure environment. Only learned parameters or gradients — not raw patient data — are shared back with a central server or aggregator, which updates the global model accordingly. This process is iterative, coordinated, and secure.
Institutions like Mayo Clinic, MD Anderson, and St. Jude hold some of the world’s most valuable and rare patient datasets — from pediatric oncology to rare genetic diseases. But these datasets are often siloed due to privacy constraints, governance requirements, or infrastructure mismatches.
Federated AI allows these institutions to collaborate on AI training and model improvement across institutions, even globally, while still keeping patient data fully local and sovereign.
Real-world benefits include:
- Stronger models trained on more diverse patient populations (racial, demographic, rare disease-specific)
- Improved generalization and robustness in AI diagnostics
- Faster research cycles without inter-institutional legal overhead
- Compliance-by-design — no raw data leaves the facility
AI Infrastructure Considerations for Life Sciences and Research Hospitals
Making federated AI work in real clinical environments isn’t just about training models — it’s about deploying them responsibly at scale, and repeatably across a fragmented healthcare infrastructure.
To achieve this, institutions must invest in secure orchestration platforms to coordinate model updates, versioning, and auditability. Techniques like differential privacy ensure that shared
model updates cannot be reverse engineered into identifiable patient data. Additionally, homomorphic encryption and secure multiparty computation (SMPC) are being increasingly integrated to protect both data-in-use and model parameters across training nodes. Standardized execution environments (via containerization, GPU isolation, etc.) ensure that models behave predictably across sites with heterogeneous infrastructure.
However, none of this matters if the underlying data architecture isn’t AI-ready. That’s where infrastructure platforms like DDN’s Infinia become essential. AI models, especially those operating in federated or multi-modal pipelines, demand high-throughput access to distributed data and they must do so across multiple protocols (POSIX, S3, GCS, HDFS) and deployment environments (on-prem, private cloud, multi-cloud). Infinia’s architecture supports these needs natively, not through bolt-on integrations, but by abstracting the storage fabric itself into a multi-protocol, flash-first layer that speaks the language of every AI pipeline.
The analogy here is not to a vendor-specific solution, but to an open-source operating layer—a kind of “Linux for medical data”—that federated learning frameworks like NVIDIA FLARE, OpenFL, and Flower can plug into. Just as AI fails without high-quality training data, it also fails without consistent, performant access to that data, regardless of where it resides. A platform like Infinia doesn’t just store the data; it orchestrates its availability, unlocking secure collaboration across research hospitals and AI centers without forcing them to conform to a single cloud model or data format.
In short, federated AI isn’t simply a software problem. It’s a systems architecture challenge. And the institutions that recognize this and invest in shared, AI-native data fabrics will be the ones to lead in diagnostic discovery, precision oncology, and global collaborative medicine.
A Blueprint for AI-Driven Healthcare Architecture
The following diagram illustrates how a federated-ready, multi-modal data architecture can be implemented in real-world clinical environments to support high-throughput AI workflows without compromising privacy or performance.
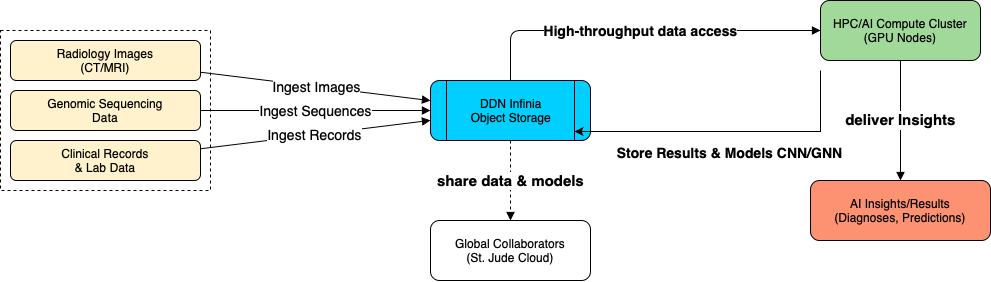
At the core is DDN Infinia Object Storage, serving as the high-performance, protocol-flexible fabric for ingesting, storing, and orchestrating access to diverse biomedical data types, including radiology images (CT/MRI), genomic sequences, and structured clinical records. Infinia ingests these datasets using native support for POSIX, S3, and GCS, enabling seamless integration with both legacy systems and next-gen pipelines.
AI models, including convolutional neural networks (CNNs) and graph neural networks (GNNs), run on connected GPU-based HPC clusters, which draw on Infinia’s high-throughput data access layer to deliver insights in real time. These models not only generate diagnostics and predictions, but also store their results back into Infinia, creating a continuous feedback loop.
Critically, this architecture supports global collaboration by enabling secure model and dataset sharing with external institutions, such as through initiatives like the St. Jude Cloud — without compromising patient sovereignty or data locality. This is how federated AI becomes operational: a shared foundation where data remains private, yet collective learning accelerates globally.
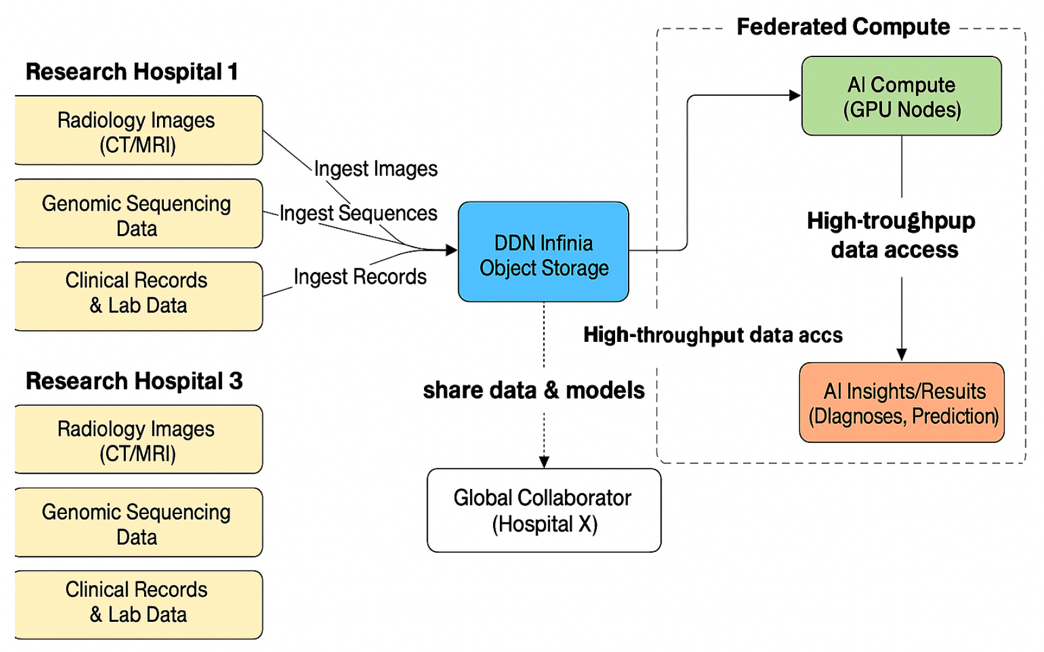
This isn’t just infrastructure — it’s a reference design for institutions committed to building the future of collaborative, AI-powered medicine. The following diagrams take the above model and show how a distributed collaborative environment may exist across multiple facilities.
Clinical Reasoning at Scale: From CNNs to GNNs
Much of the progress we’ve seen in medical AI—particularly for radiology—has been powered by convolutional neural networks. CNNs excel when applied to highly structured, low-variance datasets like chest x-rays. These images are aligned, dense, and spatially redundant. That’s the sweet spot for a CNN: a consistent field of view where learned filters can slide across every pixel, reusing and refining weight patterns to spot familiar signals—nodules, infiltrates, shadow patterns. They’re efficient, elegant, and absolutely necessary for domain-specific visual tasks that don’t require broader reasoning. But that’s also where their limitations become clear.
A chest x-ray doesn’t exist in isolation. It’s a single chapter in a longer story—a story that also includes white blood cell counts, tumor marker panels, prior diagnoses, and time-stamped progress notes. CNNs are not designed to reason over that structure. They don’t know how lab values relate to an image. They don’t track trends. They can’t natively encode the kind of mental model a clinician carries when they ask, “Why did this patient present now? What changed?” That’s where graph neural networks (GNNs) step in.
GNNs allow us to treat each of these disparate data elements—imaging features, lab values, ICD codes, genomics, encounters—as nodes in a graph. They wire these nodes together using real-world relationships: temporality, co-occurrence, correlation, and causality. During inference, message-passing steps propagate information across this graph in a way that mimics how a clinician scans a chart: finding echoes between abnormal labs and recent radiology,
contextualizing results against past medical history, triangulating meaning. It’s not a generic neural net anymore—it’s a reasoning engine.
This is exactly the architectural design I see unfolding in federated pipelines across research hospitals. I envision a local CNN performing fast, high-fidelity feature extraction on pediatric imaging. These compact feature vectors then move—not across networks, but into a local GNN trained on institution-specific context. And that GNN, in turn, contributes its updated gradients into a broader federated AI estate that spans other pediatric cancer centers. Each site retains data sovereignty. Each contributes its insights. But the intelligence is shared.
There’s no question that GNNs are less efficient than CNNs when operating in isolation. They require deeper datasets and more complex training cycles. But they offer something else: they bridge data modalities and interpret interactions. And when federated across institutions through platforms like Infinia, they enable cross-hospital reasoning without ever exposing a patient’s raw data. This architecture is not just scalable—it’s collaborative by design. CNNs remain the domain experts. But GNNs are how we turn diagnosis into understanding.
Signal Intelligence in Practice: MedGNN and Biomedical AI Inference
Our team recently worked with the MedGNN architecture, a spatiotemporal graph learning model designed to classify medical time series data. Originally developed by a multi-institutional research group — including Oxford, CUHK Shenzhen, and several Chinese universities — MedGNN represents a compelling evolution in how we think about patient-level inference from EEG signals and similar continuous monitoring data.
We deployed the model on a modest, single-GPU EC2 instance (T4, 16GB), which proved more than sufficient for both training and inference. This setup allowed us to keep costs low while testing generalizability on a real-world neurological dataset. After resolving a few minor dependency issues, we trained MedGNN on the APAVA EEG dataset — 1-second, 16-channel brainwave segments collected from 23 patients — each labeled as either healthy or diagnosed with Alzheimer’s. The dataset offered a compact but clinically relevant sandbox for evaluating how well the model could differentiate signal drift from signal meaning.
What makes MedGNN architecturally interesting is how it incorporates multiple views of the data into a unified pipeline. It doesn’t assume fixed spatial relationships between sensors but learns adaptive graph structures across time scales. It also leverages first-order difference attention to remove noise and baseline drift and combines frequency and time-domain analysis via Fourier-based convolutions. These are stitched together using a graph transformer that integrates insights from multiple resolutions before final classification.
In our experiments, MedGNN performed remarkably well, hitting nearly 99.6% accuracy on the held-out test set. More importantly, it demonstrated a strong ability to generalize without overfitting, capturing the key signal dynamics we’d expect a clinician to notice, but at a speed and scale no human could match.
While our test case involved EEG data, the architectural principles here generalize to the broader work we’re doing across institutions. When deployed within federated AI pipelines — alongside patient-specific CNNs and GNNs trained on imaging, labs, and genomics — architectures like MedGNN offer the potential to act as intelligent signal filters embedded deep within the diagnostic stack. They help compress noisy physiological data into features that matter and pass that signal into larger clinical reasoning engines downstream.
This is how we see the future: specialized models doing what they do best, stitched together within a federated infrastructure that reflects how clinicians actually think — integrative, temporal, and grounded in context.
Final Thoughts: A Shared Architecture for AI in Drug Discovery
In the coming decade, AI will not just support medical decision-making — it will shape how institutions collaborate, how diagnoses are made, and how cures are discovered. But none of this happens without the right data architecture. We can’t rely on siloed systems, single-cloud dependencies, or one-size-fits-all models. What we need is a federated, multi-modal, and secure infrastructure that allows hospitals to learn together — without giving up control. The architectural model outlined here, and validated through early experiments like MedGNN, provides a path forward. If Linux helped build a shared computing foundation for software, Infinia and federated AI can do the same for medicine.
Federated AI allows hospitals to train AI models collaboratively without moving patient data, preserving privacy and complying with HIPAA and GDPR.
Infinia provides high-performance, multi-modal AI data storage and orchestration, enabling real-time AI pipelines across hospitals and research centers.
It enables secure collaboration on rare disease and genomic datasets, improving model accuracy and research velocity without compromising data privacy.
Graph neural networks connect disparate patient data like labs, imaging, and genomics to mimic clinical reasoning and improve diagnostic accuracy.
AI-powered inference engines compress and analyze complex datasets in real time, enabling faster iteration and insight generation in drug development.



